In this month’s edition of the Honeydew Consulting Blog, we wanted to take some time to highlight some interesting and helpful build being done by one of our owner/consultants, Jack Ho. Along with UC Davis Health, Jack has developed a microbiology use-case for flagging complex multi-drug resistant organisms (MDROs) using score-based rules. MDROs are important to identify as quickly and easily as possible in order for physicians to make timely patient care decisions.
Legacy functionality for UC Davis Health’s previous Laboratory Information System (LIS) allowed for programmatic code-based logic to flag these organisms. Jack and UCDH needed to find a way to create build in Beaker to accommodate the same type of flagging. Score-based rules proved to be the way to go.
Score-based rules
Before we dive in, let’s take a moment to talk about what score-based rules are and how they help us accomplish our goal. Score-based rules are rules in which we assign a score (a number) to the outcome of the rule. Basically, if the rule meets a set of parameters, it gets the score. Score-based rules can be weighted using higher scores for certain rules/parameters based on your needs, or can be same score across all rules that meet the defined parameters. The score-based rules are then “combined” and another rule is set determining the threshold over which the score should trigger some action. In this case, the action is a flag for the organism.
Exploring MDRO build for RGNR
Epic allows for simple MDRO build that will identify the most common MDROs (Methicillin-resistant Staphylococcus aureus (MRSA) or Vancomycin-Resistant Enterococci (VRE)) based on simple organism to antibiotic combinations. Currently, it can even combine antibiotics into groups such as beta-lactams, fluoroquinolones, aminglycosides, etc. in order to assign these combinations of organisms and antibiotic groups.
But, what do you do when your MDRO protocols become more complex than what is currently in use? Explore with us below as we break down our use-case for using score-based rules to build more complex MDRO protocols for Resistant Gram Negative Rods (RGNR).
Building MDRO protocols for RGNR
Our ultimate goal is for Epic to flag an organism as “RGNR” if it is resistant* to more than two groups of antibiotics.
Some things to note: *The term resistance is used here to indicate that the organism is not sensitive to a given group of antibiotics; both intermediate or resistant results will count as “resistant.” Additionally, if any group has a reported sensitivity, that group will not count toward the score. However, if the organism shows sensitivity and resistance to the group, but the drug showing sensitivity is suppressed from the chart, then it will count toward the score.

A quick reminder to those familiar with microbiology, and a minor crash-course for those who are not…Some of the most common Gram negative groups are Acinetobacter, Burkholderia, Enterobacteriaceae, Shigella, and Stenotrophomonas. This is, by no means, an exhaustive list. Antibiotic groups we are interested in include Penicillins, Aminoglycosides, Fluoroquinolones, Cephalosporins, and Carbapenems. Again, these are just some of the more common antibiotic groups. Your microbiology and infectious disease doctors and/or departments would help drive the antibiotic groups you would need to account for in your build.
Challenges
Build such as this is not without challenges, so let’s talk about the ones that we ran into or noted so far. Two main challenges encountered were updates using cyclical logic which was removing flags incorrectly, and that the build does not work well with Bugzy/ICON.
First challenge, due to the updates of different antibiotics being reported when reclassified as a RGNR, cyclical logic can remove the flag on the isolate.
For example, if there was a drug that the isolate was sensitive to but it was only reported after RGNR-based preference reporting rules, that drug will not eliminate a group from the score-based calculation if the rule is run again (i.e. Edit/Save in Result Entry). To address this challenge, we considered using another rule property to check for the presence of the organism comment in order to prevent the rule from running on isolates that have already been flagged. Ultimately, we decided not to go that direction because it would not allow the system comment to be removed if it was applied incorrectly (due to antibiotics results being entered incorrectly).
Second challenge, Bugzy/ICON issues – due to Epic basing much of the Infection Control functionality on the MDRO organism records, workarounds are needed to identify MDROs that were flagged by this method.
Using MDRO records integrates much better throughout Epic, however as mentioned earlier, their configurability is very limited and not able to accommodate these more complex classifications. If you are not using Beaker, then MDROs are strictly the way to go as the 3rd-party LIS will take care of the MDRO assignment and interface map to a MDRO.
Let’s break down the Build
Let’s dive into pieces of the actual build. This portion is admittedly complex and may be hard to visualize. We have created images to assist, but recommend playing around with it in your test/support environment or a foundation environment, if you would like to see it all come together for yourself.
Rule Context (FCT)
First we need to make a rule context that will allow us to use our score-based rules.
- Make a copy of the Lab Result Based Context
- Update item FCT-255 to allow score-based rules
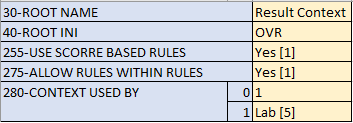
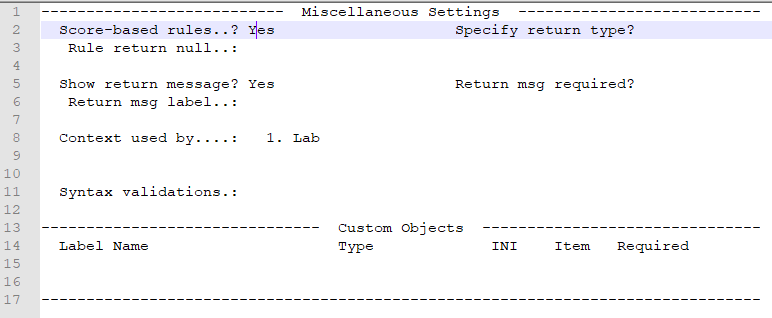
Rules (CER)
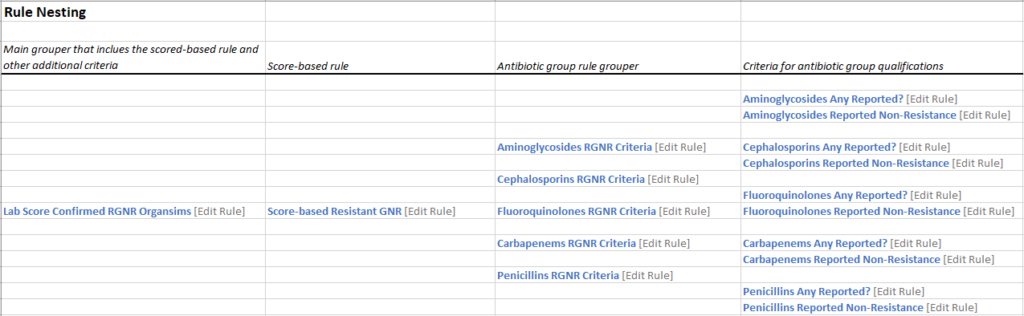
Next, we need to create the rules that make all of this work. The rules are “nested,” and we have added a visual of the nesting. We will be covering the Parent Rule, Score-based Rule, and one of the Grouper Rules. The other grouper rules work the same way with each respective drug. Please see the below images created to help illustrate the build. Please note also that this is not the recommended sequence of build. These rules build off of each other. We are presenting these in the order that they nest, not the order of build.
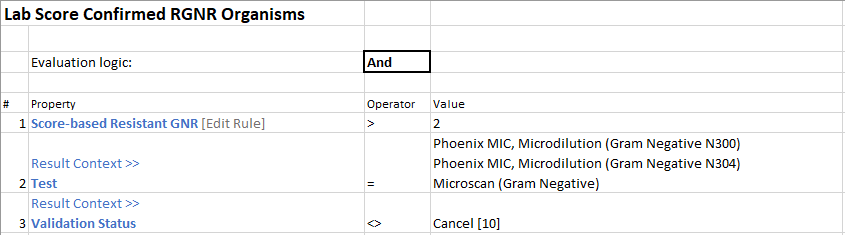
Parent Rule: This rule is used to evaluate your score-based rule. In our example, we want to evaluate for cases where our score-based rule scores higher than 2. Further qualifiers can also be added, such as test-specific or status-specific values.
Score-based Rule: This rule is used to score antibiotic group qualifications. All of the Grouper Rules (below) make up this rule. Each line scores 1 if true. The result field could be assigned different numerical values if needed to give preferential weighting to each property, but we did not for our use case.
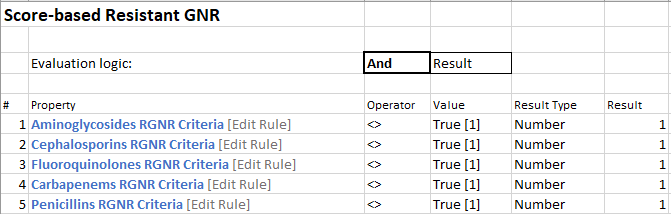
Grouper Rule: Each grouper rule is actually made up of two other “sub-” rules. Each is looking for if the “Any Reported?” rule is False OR if the “Reported Non-resistance rule” is true. There will be a grouper rule for each drug. We are only illustrating one group here (Aminoglycosides).
Grouper Rule:
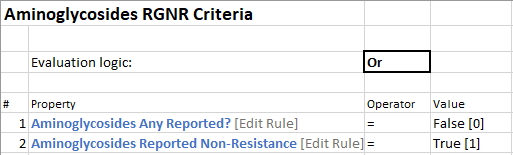
Sub-rules that make up Grouper Rule:
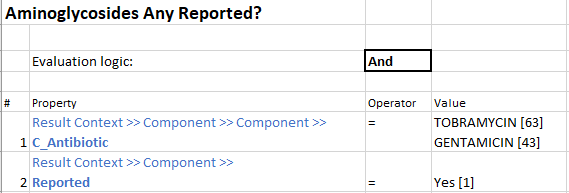
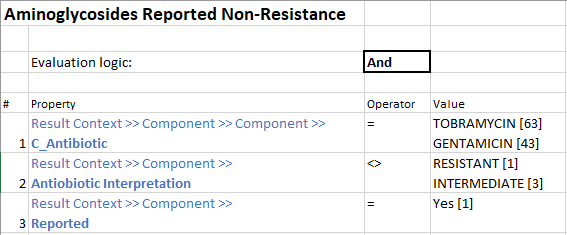
The remaining drugs’ Grouper Rules are created the same way. Thus, rolling everything up into one neat package that can help physicians make informed decisions regarding patient care.
Lab Preference Rules (LDF)
Finally, we add our CER rule to be used in the lab preference rule. Here we use the preference rules to systematically add an organism comment that we use as a flag to identify our RGNR MDRO as well as create different antibiotic reporting profiles based on this MDRO classification in combination with the organism group or genus.
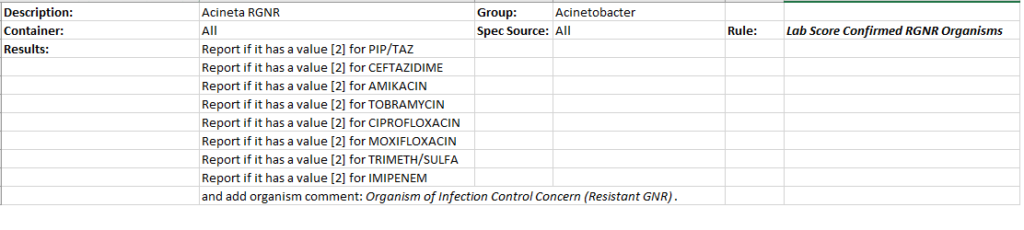
Thank you, Jack and UC Davis Health for sharing this elegant and important build!
We recognize that this is intricate and in-depth build. If it seems like something that your organization could benefit from and you would like further information, please feel free to reach out to us at [email protected].
